High Finesse is a German optics company that makes high-quality spectrometers and wavelength meters. I’ve written a python interface to their provided wlmData driver in order to grab spectrum data off a LSA UV Vis 2, however, it is likely to work with other spectrometers and wavelength meters that use the same library. Unfortunately, at this time High Finesse only provides windows drivers.
Installation
To install this library, use:
git clone https://github.com/CatherineH/pyHighFinesse
cd pyHighFinesse
python setup.py install
The project will soon be available on pypi as pyHighFinesse.
Example Script
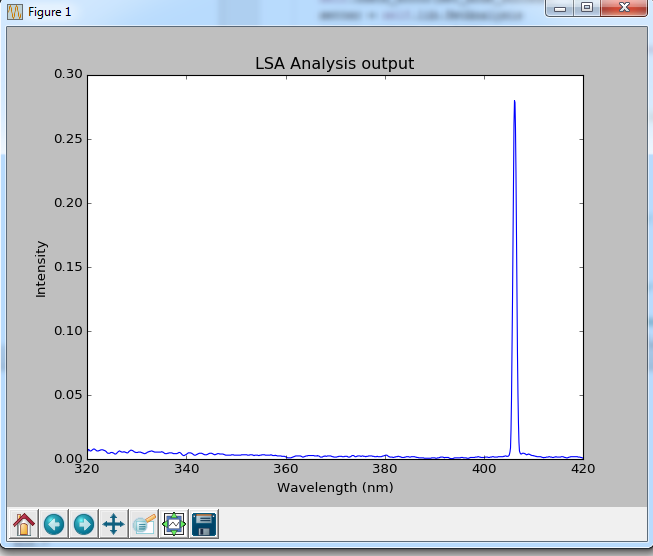
The following minimal script grabs and plots the spectrum dataframe
from lsa import Spectrometer
import matplotlib.pyplot as plt
reading = Spectrometer()
spectrumdata = reading.spectrum
spectrum = spectrumdata.set_index('wavelength')
_ax = spectrum.plot()
_ax.legend_.remove()
_ax.set_title('LSA Analysis output')
_ax.set_xlabel('Wavelength (nm)')
_ax.set_ylabel('Intensity')
plt.show()
The WLM and LSA
High Finesse provides a dynamic library called wlmData.dll, which is installed to the system32 directory. They also provide a C header called wlmData.h which contains additional constants useful to the interface such as error codes and options.
The LSA is a GUI that also a server process that communicates with the instrument. The python interface communicates and shares memory with the LSA through the dynamic library. The python interface needs to start up the LSA in order to get data off the spectrometer.
Parsing the Header file
The header file contains a lot of useful information about error codes, return types and options. In future, the function return type could be used to automatically populate the ctypes restype value, but for now only error codes and options are red in.
The constants section of the header file is deliminated by a line-long comment that contains the word “Constants”, so parsing begins after that line is encountered
begin_read = False
for line in f_in.readlines():
if line.find("Constants") > 0:
begin_read = True
Next, if a line contains the keywords const int, the line probably contains a useful value to parse. The keywords, whitespace characters and semicolon are removed, and the string is split around the equals sign
if begin_read and line.find("const int") > 0:
values = line.split("const int")[1].replace(";", "") \
.replace("\t", "").replace("\n", "") \
.split("//")[0].split(" = ")
Header values are added as attributes to the Spectrometer object. The values of the integer constants are one of three things: a number, a string representing a previously declared integer constant, or an expression adding previous constants and numbers. Since there is no built-in function to check whether a string can be parsed as an integer, this parsing is done as a nested try/catch block. First the string is parsed as an int; if that fails, it is split around the plus sign. (The header expressions currently only contain additions). Then, there must be a second integer check, and if that fails, it assumes that the string is a value that python has previously parsed in.
try:
# first, attempt to parse as int
setattr(self, values[0], int(values[1], 0))
except ValueError:
parts = values[1].split(" + ")
# if that fails, parse as a value
value = 0
for part in parts:
try:
value += int(part, 0)
except ValueError:
value += getattr(self, part)
setattr(self, values[0], value)
Errors should be added to their corresponding sets for future error checking. Constants starting with Err occur when measurement values or settings are read from the LSA; constants starting with ResErr occur when settings are changed in the LSA. The list of ResErr errors contains an error 0 for NoErr to confirm that the setting was changed, this error is purposely excluded fro m the list of errors.
if values[0].find("Err") == 0:
if 'read' not in self.errors_list.keys():
self.errors_list['read'] = []
self.errors_list['read'].append(values[0])
if values[0].find("ResERR") == 0:
if 'set' not in self.errors_list.keys():
self.errors_list['set'] = []
# no not append the "NoErr" Error
if getattr(self, values[0]) != 0:
self.errors_list['set'].append(values[0])
Finally, the wavelength ranges are also defined in the header, however these do not appear to correlate with the actual values in the LSA. For the moment, this is hard-coded for the observed values. If you have a High Finesse device other than an LSA, please let me know what these ranges are for you.
self.wavelength_ranges = [(0, (190, 260)), (1, (250, 330)),
(2, (320, 420))]
Accessing Memory
The functions in the WLM dynamic library will either return a value, such as a short, long or double, or an address in memory. The return type must be specified in order for the output to be read correctly.
For example, c_double must be specified when reading floating point numbers, such as temperature or wavelength:
get_wavelength = self.lib.GetWavelength
get_wavelength.restype = c_double
wavelength = get_wavelength(0)
Other entries, such as GetPattern or GetAnalysis will return an address in memory of an array of numbers. The number of numbers can be queried using the GetAnalysisItemCount, the size of the numbers in bytes can be queried using GetAnalysisItemSize: 2 for c_short 4 for c_long and 8 for c_double. To remove redundant code, all three values are queried using the same lines of code by looping over the parameters for each of the x and y axes of the analysis pattern:
results = {}
parts = [{'wavelength': 'X', 'intensity': 'Y'},
{'size': 'ItemSize', 'count': 'ItemCount', 'address': ''}]
for axis in parts[0]:
results[axis] = {}
for value in parts[1]:
getter = getattr(self.lib, 'GetAnalysis'+parts[1][value])
getter.restype = c_long
component_arg = getattr(self, 'cSignalAnalysis'+parts[0][axis])
results[axis][value] = getter(component_arg)
These values can then be read in using the cast function:
DATATYPE_MAP = {2: c_int, 4: c_long, 8: c_double}
...
memory_values = {}
for axis in parts[0]:
access_size = DATATYPE_MAP[results[axis]['size']]*results[axis]['count']
memory_values[axis] = cast(results[axis]['address'],
POINTER(access_size))
Finally, the individual numbers in the array can be accessed using the contents property. For ease of use in analysis, the values are then stored in a pandas dataframe:
spectrum_list = []
for i in range(0, results['wavelength']['count']):
spectrum_list.append({'wavelength': memory_values['wavelength'].contents[i],
'intensity': memory_values['intensity'].contents[i]})
spectrum = DataFrame(spectrum_list)
LSA Oddities
The WLM driver occasionally behaves in ways that contradict the user manual. For example:
- Hiding the WLM control does not also hide the long term analysis window
- The wavelength ranges provided in the header do not correspond to the actual wavelength ranges on the instrument
- The parameters for wide and precise spectrometer analysis appear to be reversed.
It is likely that other High Finesse devices also exhibit odd behavior, however this module was currently tested with an LSA UV Vis 2. If this module results in odd behavior on your device, please let me know by submitting an issue on github.